When wandering around the department, I am struck by how few crystallographers use multiple contoured electron density maps* whilst building. I really don’t understand this: YOU’RE THROWING AWAY INFORMATION PEOPLE!
Even at moderate resolution, the information gained can be invaluable:
- Precisely locating a heavy atom in a big blob of electron density. The Atom will sit at the highest point of the map. IF you are using multiple contours, this will be obvious! (figure 1)
- Resolving His/Asn/Gln sidechain flips. A bit more prone to noise here – but in terms of electrons, O > N > C. You can easily decide which way around Asn and Gln side chains should point, and often get some help with His side chains as well. (figure 2)

Figure 1a: THAR SHE BLOWS!
This calcium ion sits RIGHT on the peak in the electron density map. No ambiguity where it lies. A single map contoured at 1 sigma is not helpful.

Figure 1b: Nope. Not helpful.

Figure 2: Gln sidechain flips, made easy…
The increased electron density on the right hand side here indicate that this Gln residue is the correct way around. Again, a single map contoured at 1 sigma is no use here.
I realise that there are other ways to achieve what I have described, but when you are building your models, saving time and making things easier is hugely helpful. I find using multi-contouring incredibly helpful.
* multi-contours shown here are made using the “Multi-chicken” command in COOT (extensions>maps>multi-chicken). Multi-chicken creates 10 maps contoured at 1,1.5,2, 2.5, etc sigma. I find the default setting is a tad dark so I use “brighten maps” (extensions>maps>brighten maps) a couple of times to sort that out. I then contour the original map at 0.7sigma (depending upon noise) and make it really deep purple.
All screenshots made with COOT.
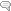
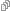

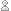 Posted by xtaldave
Posted by xtaldave 

